Over the last 70 years, antibiotics and similar drugs have greatly reduced illness and death from infectious diseases. Many drugs, however, no longer work as well as they once did because the infectious organisms they are designed to kill have adapted to them.
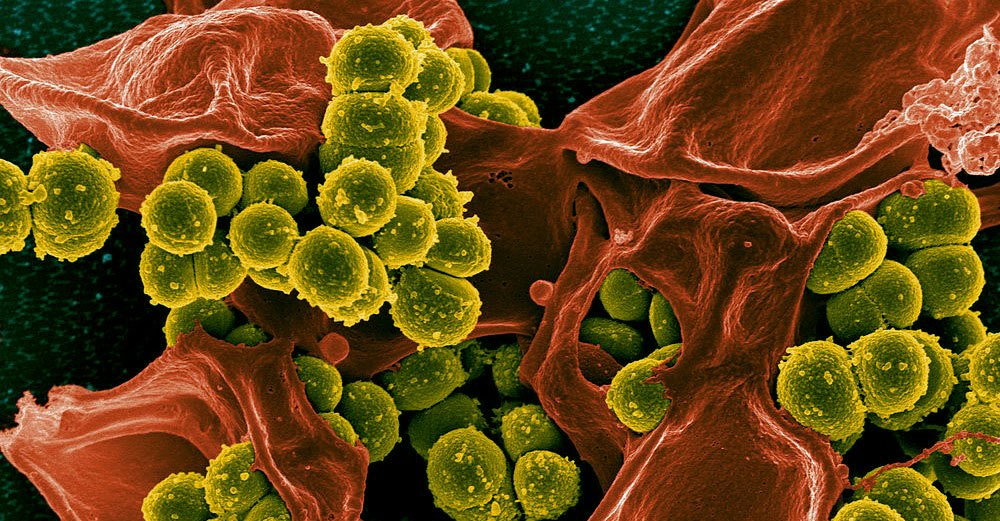
So-called MRSA infections, which are prevalent in hospitals and elsewhere, are one example of the problem. MRSA, or methicillin-resistant Staphylococcus aureus, is a type of staphylococcus or “staph” bacterium that is resistant to many antibiotics. Typically found in skin infections, MRSA caught in medical facilities can bring on life-threatening blood and surgical site infections, as well as pneumonia.
The growing public health threat of MRSA is spurring research to develop new treatments. To advance the work of SUNY researchers in this arena, the SUNY Health Network of Excellence is funding a project led by Paul Agris, director of The RNA Institute at the University at Albany. As its name indicates, The RNA Institute focuses on the RNA molecule and ways to harness its features to diagnose and treat disease.
“While DNA contains the multi-step instructions for all proteins, RNA builds proteins using these instructions. RNA controls which proteins are built and their quantities,” explains Agris. “RNA, as a novel therapeutic target for development of new classes of drugs, has the potential to target any step of protein building, including preventing the production of bacterial proteins needed to cause infection.”
In the 1940s, medical treatment for Staphylococcus aureus (S. aureus) infections, commonly known as “staph,” became routine and successful with the discovery and introduction of antibiotic medicine, such as penicillin. In the late 1940s and the 1950s, S. aureus developed resistance to penicillin. Methicillin, a form of penicillin, was introduced to counter the increasing problem of penicillin-resistant S. aureus, but, in 1961, scientists identified the first strains of S. aureus bacteria that had become resistant to methicillin treatment, now known as MRSA. S. aureus continues to evolve and now shows resistance to numerous antibiotics.
“To counter the relentlessly developing pandemic of these multidrug-resistant pathogens, we are taking a new approach that focuses on a family of closely related RNAs as targets for antimicrobial agents,” says Kyla Frohlich, the lead research scientist with Agris at UAlbany. “Our goal is to validate novel drug candidates as a viable new class of antibiotics that kill the MRSA bacterium, but display little-to-no toxicity to human cells.”
“These RNA targets were chosen because they are required for the growth of a large number of bacteria that cause infections, but are not present in humans. This means we could target bacterial RNA and effectively treat a variety of multi-drug-resistant bacterial infections without affecting the human RNA,” explains Frohlich.
Using advanced computer algorithms, 250,000 small molecules were screened for their ability to bind the bacterial RNA target. “We then used this information to conduct experiments in the lab that have identified a potential new antibiotic that kills a wide range of multi-drug-resistant bacteria, including MRSA, and have little to no effect on human cells,” says Frohlich. Patent applications are filed by SUNY RF.
Collaborating with Agris and Frohlich are UAlbany researchers Alan Chen, assistant professor, Department of Chemistry and The RNA Institute, and Kathleen McDonough, research scientist, Wadsworth Center, New York State Department of Health and UAlbany’s School of Public Health; Douglas Kitchen of Albany Molecular Research Inc.; and Paul Kostyniak and Patricia Masso-Welch of the University at Buffalo.
The team aims to develop new approaches to combating antibiotic-resistant bacteria, and, says Agris, “we plan to use our findings to support a multi-investigator proposal for funding to the National Institutes of Health.”
In the future, the threat of these dangerous infections may be classified as a thing of the past.




Hope you can make a great progress in future!
Really a nice post, and thanks for sharing with us.